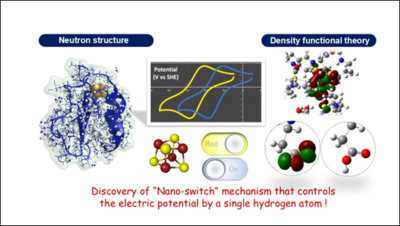
Elucidating the redox potential regulation mechanism common to all living organisms by an "electron carrier" protein for energy acquisition- Discovery of a "nano-switch mechanism" controlled by a single hydrogen atom -
Osaka University
Ibaraki University
University of Miyazaki
Tokyo University of Pharmacy and Life Sciences
Kurume University
Ibaraki Prefecture
J-PARC Center
Comprehensive Research Organization for Science and Society (CROSS)
Japan Synchrotron Radiation Research Institute(JASRI)
A research group led by Professor Yasutaka Kitagawa of Osaka University, Professor Kei Wada of Miyazaki University, and Professor Masaki Unno of Ibaraki University (in collaboration with researchers from Tokyo University of Pharmacy and Life Sciences, Kurume University, CROSS, and JASRI) has revealed a mechanism for controlling the potential of an "electron carrier" protein in the redox reaction that all organisms need to obtain energy. Based on experiments using the Ibaraki Biological Crystal Diffractometer (iBIX) at the Materials and Life Science Experimental Facility (MLF) in the Japan Proton Accelerator Research Complex (J-PARC), the precise 3D structure of the protein including hydrogen atoms was determined, and theoretical calculations using this data visualized the electronic structure of the iron-sulfur cluster. As the results, it was revealed, for the first time, that the electric potential of the iron-sulfur cluster changes dramatically depending on the presence or absence of a single hydrogen atom at an amino acid side chain, a so-called "nano-switch" mechanism.
This study was published in the online edition of the international scientific journal eLife*1 on November 15, 2024 (Reviewed Preprint).
Points
∗ Most reactions in living organisms involve the "electrons" transfer, which is called redox reaction. For example, respiration and photosynthesis can be classified as redox reactions. Some proteins that assist in the electron transfer contain irons and sulfurs.
∗ Ferredoxin is a small protein that holds iron-sulfur clusters inside it and is known as the "electron carrier" in living organisms. Ferredoxin is a universal protein that is thought to be present in almost all living organisms, however, the mechanism by which ferredoxin stably carries electrons has remained a mystery to date.
∗ In this study, we have succeeded in determining the precise three-dimensional structure of a ferredoxin at the hydrogen atomic level in experiments using a neutron beam. Visualizing hydrogen atoms in protein molecules using neutrons is extremely difficult, and only less than 0.2% of the entire protein three-dimensional structure database (Protein Data Bank; PDB) has been reported.
∗ Theoretical calculations using experimental geometry including hydrogen atoms were performed to elucidate the electronic structure of the iron-sulfur cluster in the ferredoxin. As a result, it was revealed, for the first time, that an amino acid residue (aspartic acid 64) located far from the iron-sulfur cluster has a significant effect on probability of electron transfer in the iron-sulfur cluster, and plays a role like a switch that controls the electron transfer in ferredoxin. Furthermore, it was shown that the mechanism is universal in various organisms.
∗ The results will not only deepen our scientific understanding of biological reactions but also provide a major clue to the future development of ultra-sensitive sensors for oxygen and nitric oxide and novel drugs.
Background
In living organisms, electrons are constantly being transferred between substances. Donating electrons to a substance is called "reduction"; drawing electrons from a substance is called "oxidation". These repeated reactions are called "redox reactions". Respiration and photosynthesis are typical examples of redox reactions in living organisms.
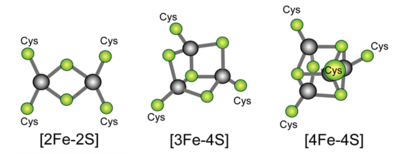
In living organisms, various proteins assist in redox reactions, some of which contain clusters of iron and sulfur (iron-sulfur clusters), and these iron-sulfur clusters play important functions in the transfer of electrons between proteins. Ferredoxin, a small protein containing iron-sulfur clusters, is thought to be present in almost all living organisms and is a typical example of an "electron carrier". The history of ferredoxin research is old; it began 60 years ago. Various types of ferredoxins with various iron-sulfur clusters which contain different numbers of constituent irons and sulfurs have been discovered (Figure 1).
Figure 1. Examples of typical iron-sulfur clusters. The yellow-green spheres are sulfurs, and the gray spheres are irons. There are cases in which some of these are present in combination in a single protein molecule. The letters Cys represent sulfurs of cysteines in the protein.
While water flows from high to low by nature, electrons flow from low to high potential energy ( "potential" or "electrostatic potential" ). The potentials (redox potentials) of ferredoxins, which have various types of iron-sulfur clusters, are diverse and wide-ranging. Ferredoxins raise and lower the redox potentials of their iron-sulfur clusters like an elevator, sometimes giving electrons to other proteins and sometimes withdrawing them from others. However, how this is controlled remains unclear.
Although theoretical calculations based on a method called "density functional theory"*2 can be used to study the electronic structure*3 of ferredoxin, it requires an accurate 3D structure of ferredoxin including the hydrogen atoms for accurate calculations. However, since it is extremely difficult to determine the position of hydrogen atoms in a protein molecule, theoretical calculations for most proteins, including ferredoxin, have conventionally used "assumed" positions of hydrogen atoms. If the assumed positions of the hydrogen atoms differ from the exact ones, the most fundamental assumptions of the theoretical calculations would break down and the obtained conclusions would be meaningless. The research group, therefore, set out to experimentally determine the positions of hydrogen atoms in ferredoxin, and to elucidate the electronic structure of the iron-sulfur cluster based on experimental facts.
Research Methods
Currently, X-ray crystallography (Nobel Prize in Chemistry 1962) and cryo-electron microscopy (Nobel Prize in Chemistry 2017) are commonly used to analyze the 3D structure of proteins with resolutions at the atomic levels, however, these methods are not suitable for identifying the smallest hydrogen atom. In addition, even the protein structure prediction algorithm (AlphaFold), which won this year's (2024) Nobel Prize in Chemistry, cannot predict the exact locations of hydrogen atoms to date. Therefore, in this study, we used neutron crystallography*4, which can identify hydrogen atoms with the same degree of clarity as other atoms in proteins.
In this research, after overcoming the high hurdle of growing very large ferredoxin crystals, we patiently collected data using neutrons at the Ibaraki Biological Crystal Diffractometer (iBIX) *5 in the Materials and Life Science Experimental Facility (MLF) at the Japan Proton Accelerator Research Complex (J-PARC) *6 in Tokai-mura, Ibaraki Prefecture The data was collected using neutrons.
Furthermore, using the experimentally determined precise structure, the electrons in the iron-sulfur cluster were investigated by theoretical calculations based on quantum mechanics and quantum chemistry (density functional theory). The results obtained were verified by using various mutants of ferredoxin, in which one amino acid in ferredoxin was altered by gene manipulation, as a sample, and by conducting experiments in a chamber where oxygen was excluded to the utmost limit to prevent oxidation of the iron-sulfur clusters by air.
Results
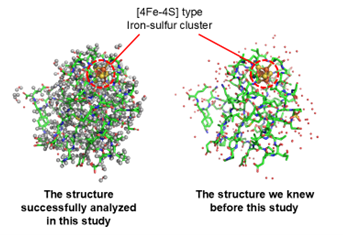
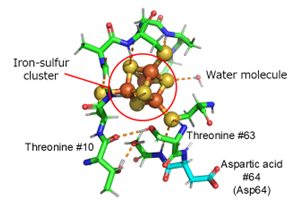
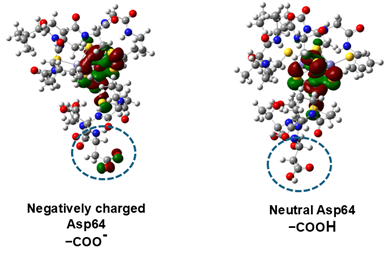
In this study, the 3D structure of ferredoxin, which has four iron and four sulfur clusters in the molecule ([4Fe-4S]-type clusters; Figure 1), was determined by neutron crystallography, and the exact positions of atoms including hydrogen around the iron-sulfur cluster were experimentally determined (Figure 2). The actual positions of hydrogen atoms around the iron-sulfur cluster were found to be different from the previously predicted position (Figure 3). Based on the exact positions of the hydrogen atoms, the electronic structure around the iron-sulfur cluster was calculated theoretically, and it was found, for the first time, that electrons originating from the iron-sulfur cluster are distributed not only around the iron-sulfur cluster but also to aspartic acid 64 (Asp64) at a distance of more than 1 nm (nanometer = 0.000001 mm) away from the cluster. (Figure 3 and Figure 4). (1 nm is too long a distance in a protein molecule for direct interaction.) Interestingly, this electron distribution was observed only in the absence of a hydrogen atom (-COO-) in the side chain (carboxy group: -COOH) of Asp64, while in the presence of a hydrogen atom (-COOH), the electrons were distributed only around the iron-sulfur cluster (Figure 4). In fact, by measuring the rate at which the iron-sulfur cluster is oxidized and the redox potential, we proved that Asp64 has a significant effect on the reactivity of the iron-sulfur cluster. Although ferredoxin contains multiple aspartic acids, only Asp64 showed such a phenomenon. Furthermore, in ferredoxins from various microorganisms, aspartic acid residues in similar 3D positions were found to affect the electronic structure of the iron-sulfur cluster.
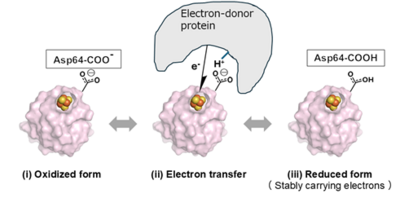
In this study, we elucidate for the first time in the world the existence of a "nano-switch mechanism" in which the presence or absence of a single hydrogen atom in the aspartic acid side chain changes the electronic state of the iron-sulfur cluster (Figure 5). This nano-switch mechanism has also been shown to be conserved in archaea and is believed to be widely used in the biological world.
Figure 2. Left: The overall structure of [4Fe-4S]-type ferredoxin containing hydrogen atoms, which was successfully analyzed using neutrons in this study. Hydrogen atoms are highlighted by gray spheres. Right: An X-ray image of the overall structure of the same ferredoxin that was previously known.
Figure 3. Structure around the iron-sulfur cluster. Hydrogen bonds are indicated by dotted lines. For example, threonine 63 was initially thought to be hydrogen bonded to the iron-sulfur cluster (circled in red), however, neutron crystallography showed that the hydrogen atom of its -OH was in the other direction, forming hydrogen bonds with the main chain of threonine 10.
Figure 4. Structure around the iron-sulfur cluster. Hydrogen bonds are indicated by dotted lines. For example, threonine 63 was initially thought to be hydrogen bonded to the iron-sulfur cluster (circled in red), however, neutron crystallography showed that the hydrogen atom of its -OH was in the other direction, forming hydrogen bonds with the main chain of threonine 10.
Figure 5. A schematic drawing of the electron transfer mechanism by ferredoxin that revealed in this study.
Future Expectations
Iron-sulfur clusters in protein molecules are involved in various reactions that play fundamental roles in biological activities. The present redox potential control switch mechanism can be applied to the control of those reactions. For example, in proteins that detect oxygen (O2) and nitric oxide (NO) in vivo, it is the iron-sulfur cluster that detects very small amounts of gases. Also, in many microorganisms, including pathogenic bacteria, [4Fe-4S]-type iron-sulfur clusters in proteins play essential roles in energy acquisition. More recently, ferredoxin and iron-sulfur clusters have been found to play important roles in cancer cells. The findings of this research will not only deepen our scientific understanding of biological reactions but also provide a major clue for the future development of ultra-sensitive sensors of O2 and NO and novel drugs (e.g., anti-cancer drugs, antibiotics against pathogens, etc.).
Terminologies
*1: eLife
eLife is a non-profit, open-access journal launched in 2012 for biomedical and life sciences. Although relatively new, it is already ranked #5 in the field of Biology and Biochemistry. https://research.com/journal/elife
*2: Density Functional Theory Method
The DFT (Density Functional Theory) method is widely known as a method that calculates energy from the electron density of atoms and molecules. It can be applied to molecules with large sizes such as proteins because it can estimate molecular energies accurately with a relatively smaller computational costs. Since accurate structural data of molecules are required for more reliable calculations, it is very important to obtain the coordinates of hydrogen atoms experimentally.
*3: electronic state
The term "electronic state" refers to the distribution and energy of electrons in a material. The function expressing distribution of each electron is called the molecular orbital, which can be used to explain the chemical bonds between atoms and, in turn, the properties of molecules.
*4: neutron crystallography
An analytical method to obtain the 3D structure of molecules in a crystal by irradiating neutrons into the crystal and measuring the diffracting intensity. It is like X-ray crystallography but allows detailed observation of hydrogen atoms (or hydrogen ions = protons) due to neutrons interacting with atomic nuclei. Since X-rays are scattered by electrons, the scattering from a hydrogen atom containing only one electron is very weak and is not suitable for identifying the hydrogen atoms in the molecule. However, whereas protein X-ray crystallography can be performed even with small crystals, very large crystals are required for protein neutron crystallography. Cryo-EM can identify hydrogen atoms more easily than X-ray crystallography; however, it is currently not applicable to small proteins such as ferredoxin. Neutron crystallography was the best way to reveal the structure of this ferredoxin at the hydrogen atom level, although it is very difficult to grow large crystals of proteins.
*5: Ibaraki Biological Crystal Diffractometer (iBIX)
It is the world's highest-level single-crystal diffractometer for high-resolution protein crystallography using the powerful pulsed neutron source of J-PARC at one of the two neutron beamlines installed at MLF by Ibaraki Prefecture.
*6: Japan Proton Accelerator Research Complex (J-PARC)
J-PARC is the generic name for the world's largest proton accelerator and experimental facilities with the world's highest beam intensity, jointly constructed by the Japan Atomic Energy Agency (JAEA) and the High Energy Accelerator Research Organization (KEK) in Tokai-mura, Ibaraki Prefecture, Japan. Using secondary particles such as neutrons, muons, mesons, and neutrinos that are produced when accelerated protons collide with a nuclear target, cutting-edge academic research and industrial applications in materials and life sciences, nuclear and particle physics, etc. At the Materials and Life Science Experimental Facility (MLF) within J-PARC, experiments can be conducted using the world's highest performance pulsed neutron and muon beams.
Research Funds
These results were obtained through the following research projects and financial supports.
● Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Transformative Research Areas (A), and Japan Society for the Promotion of Science (JSPS), Grant in Aid for Scientific Researches (B) and (C)
● Research Grant from Takeda Science Foundation, Enzyme Research Grant from the Japan Foundation for Applied Enzymology, and Ibaraki Prefecture Leading Research Project Fund (Sendo-kenkyu)
Paper Information
| Title | Protonation/deprotonation-driven switch for the redox stability of low potential [4Fe-4S] ferredoxin |
|---|---|
| Authors | Kei Wada*, Kenji Kobayashi†, Iori Era†, Yusuke Isobe, Taigo Kamimura, Masaki Marukawa, Takayuki Nagae, Kazuki Honjo, Noriko Kaseda, Yumiko Motoyama, Kengo Inoue, Masakazu Sugishima, Katsuhiro Kusaka, Naomine Yano, Keiichi Fukuyama, Masaki Mishima, Yasutaka Kitagawa*, Masaki Unno* *corresponding authors †equally contributed |
| Journal | eLife |
| Publish date | Nov. 15th, 2024 (Reviewed Preprint) |
| DOI | 10.7554/eLife.102506 |
Researchers Information
Yasutaka Kitagawa
Graduate School of Engineering Science, Osaka University
Kei Wada
Department of Medical Sciences, University of Miyazaki
Masaki Unno
Graduate School of Science and Engineering, Ibaraki University
Masaki Mishima
Department of Molecular Biophysics, Tokyo University of Pharmacy and Life Sciences
Masakazu Sugishima
Department of Medical Biochemistry, Kurume University School of Medicine
Katsuhiro Kusaka
Neutron Science and 20 Technology Center, Comprehensive Research Organization for Science and Society (CROSS)
Naomine Yano
Structural Biology Division, Japan Synchrotron Radiation Research Institute (JASRI)